Yuemin Liu

Lecturer
Education
- Sc. Analytical Chemistry, Shanxi University, 1985
- M.S. Organic/Radiochemistry, China Institute of Atomic Energy, 1991
- Ph.D. Computational Chemistry, University of Toledo, 2005
Research Interests
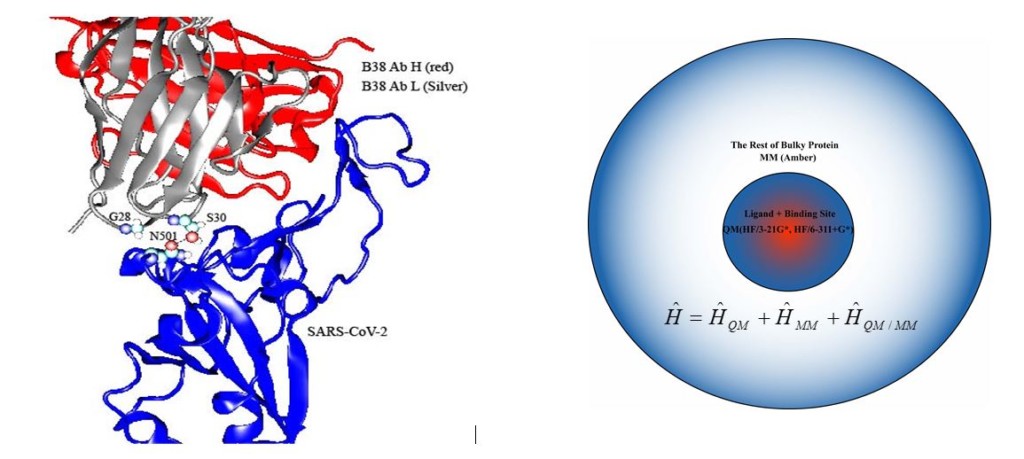
Understanding intermolecular interactions are essential to studies ranging from basic chemical reactions to complicated cell activities, thus provide insights into catalytic mechanism and molecular/drug design. Crystallography represents the most direct experimental approach to reveal wide variety of intermolecular interactions. However, the intrinsic limitations of crystallography are the missing of dynamics attributes and low resolutions in intermolecular interactions. Computational simulations proved a critical approach to mitigate above limitations of crystallography, thus play an important role in addressing diversity of intermolecular interactions. Hybrid quantum mechanics/molecular mechanics (QM/MM) method represents the best combination of two worlds of quantum mechanics and molecular mechanics. The core region of the system in which the targeted chemical process occurs can be considered at higher level quantum chemistry methods, while the larger remainder of the system is treated with a relatively less expensive molecular mechanics force field. In this work we are studying methods to decipher intermolecular interaction between S protein of SARS-CoV-2 and antibody B38 using QM or QM/MM simulations as shown in the figure below. Density Function Theory (DFT) simulation is the standard tool for studying organometallic systems. Hybrid DFT methods contain a certain portion of Hartree-Fock (HF) exact exchange, and provide reasonable treatments of dispersion forces at lower computational expenses compared to the ab-intio methods. It is very meaningful to evaluate how hybrid DFT methods behave based on benchmark Moller-Plesset and Couple Cluster theory calculations in the simulation of highly-correlated organometallic systems.
Teaching Interests
General, organic, physical chemistry, and biochemistry
Professional Activities
Member of graduate defense committee 2011-2020, Outreach activities and scholarships committee 2021-2022, and Seminar committee 2021-2022.
Selected Research Publications
- Yuemin Liu, Thomas Junk. In-depth Normal Mode Analysis of Seven Prominent Raman Bands of TNT at Bond Level from Isotopic Shifts of TNT-d5.1143, (2017) 438–443.
- Yuemin Liu, Richard Perkins, Yucheng Liu, Nianfeng Tzeng. Normal Mode and Experimental Analysis of TNT Raman Spectrum. 1133, (2017) 217-225.
- Yuemin Liu, Yucheng Liu, Siva Murru, Nianfeng Tzeng, Radhey S. Srivastava. Quantum Mechanics Study of Repulsive π-π Interaction and Flexibility of Phenyl Moiety in the Iron Azodioxide Complex. 1097 (2015) 226–230.
- Yuemin Liu, Thomas Junk, Yucheng Liu, Nianfeng. Tzeng and Richard Perkins. Benchmarking Quantum Mechanical Calculations with Experimental NMR Chemical Shifts of 2-HADNT. Journal of Molecular Structure. 1086 (2015) 43-48.
- Yuemin Liu, Yucheng Liu, August A. Gallo, Katherine D. Knierim, Eric R. Taylor, Nianfeng Tzeng. Performances of DFT methods implemented in G09 for simulations of dispersion-dominated CH-π Interaction in ligand–protein complex: A case study with glycerol-GDH. Journal of Molecular Structure. 1084 (2015) 223-228.
- Salah S. Massoud, Eric Taylor, Yuemin Liu, Janusz Grebowicz, Ramon Vicente, Roger Lalancette, Udai Mukhopadhyay, Ivan Bernal, and Steven F. Watkins. Synthesis, Structure, Thermal, Magnetic Properties and Quantum Mechanical Calculations of Bridged [Bis(di(2-pyridylmethyl)amine)-(μ2-1,2-bis(4-pyridyl)ethane)-tetraperchlorato-dicopper(II)] Dihydrate Complex (I). CrystEngComm 2014,16, 175-183.
- Yuemin Liu, August A. Gallo, Jan Florian, Wu Xu, and Rakesh Bajpai. CH···π Interactions Do Not Contribute to Hydrogen Transfer Catalysis by Glycerol Dehydratase. J. Phys. Chem. ((2011) 115(41); 11162-11166.
- Yuemin Liu, August A. Gallo, Rakesh K. Bajpai, Andrei Chistoserdov, Andrew T. Nelson, Leah N. Segura and Wu Xu. The diversity and molecular modeling analysis of B12-dependent and B12-independent glycerol dehydratases. Int. J. Bioinformatics Research and Applications. (2010) 6(5), 484-507.
- Yuemin Liu, August A. Gallo, Jan Florian, Yen-Shan Liu, Sandeep Mora and Wu Xu. QM/MM (ONIOM) Study of Glycerol Binding and Hydrogen Abstraction by the Coenzyme B12-Independent Dehydratase. J. Phys. Chem. (2010) 114(16); 5497-5502.
- Yuemin Liu, Di Pan Susan L. Bellis, Yuhua Song. Effect of Glycosylation and Variant Sialylation on β1 Integrin Structure and Function: A Molecular Dynamics Study. Proteins: Structure, Function, Bioinformatics. 73: 989-1000 (2008).